New Customers Sepcial: Get 15% off, use the code "NEW15" at checkout. Shop Now!
Unlock Molecular Insights with Custom FISH Probes
At TridixBio, our Fluorescence In Situ Hybridization (FISH) probe services are designed to empower your research with precision and clarity. From custom probe design to advanced imaging support, wew provide end-toend solutions tailored to your unique molecular discovery goals.
Our FISH Probe Solutions

Custom Probe Design: Tailored probes crafted to target your specific gene sequences for unparalleled specificity.

High-Performance Fluorescent Labeling: A wide range of premium dyes to ensure vibrant, clear signals for accurate detection.

Comprehensive Application Support: Expert guidance on hybridization protocols, imaging techniques, and data analysis.

Versatile Assay Compatibility: Optimized for gene expression studies, chromsomal abnormality detection, RNA localization, drug screening, and more.Comprehensive Application Support: Expert guidance on hybridization protocols, imaging techniques, and data analysis.
Why Choose TriDix Bio?
Precision & Quality
Meticulously designed probes ensure reliable, reproducible results.
End-to-End Expertise
From probe synthesis to data interpretation, we support every step of your workflow.
Customized Solutions
Flexible services and expert support tailored to your research objectives, regardless of complexity.
Accelerated Discovery
High-quality probes and short turnaround time to fast-track your molecular insights.
Service Flowchart for FISH Probes

FISH Probe Imaging Showcase
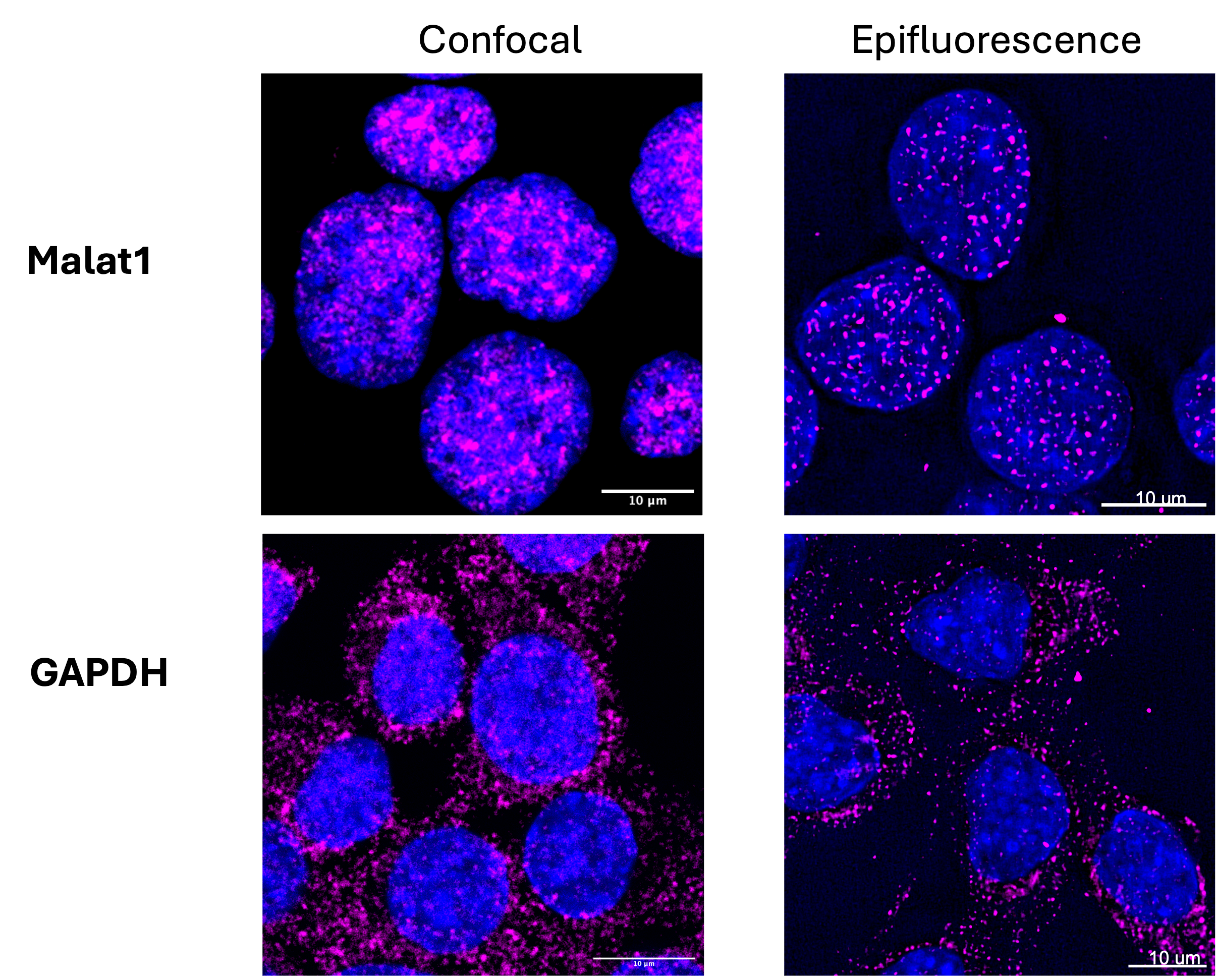
Figure 1, FISH signal visualization with confocal and conventional fluorescent microscopes: Neuro-2a cells were stained for mouse MALAT1 lncRNA(top) and GAPDH mRNA using corresponding FISH probes, labeled with ATTO647N dye. Nuclei were counterstained with DAPI. Imaging was conducted either using a Leica TCS-SP8 confocal microscope with a 63x oil immersion objective (Left) or Leica TIRF/D STORM/THUNDER (widefield) with a 63x objective with oil (Right). Scale bar: 10 µm
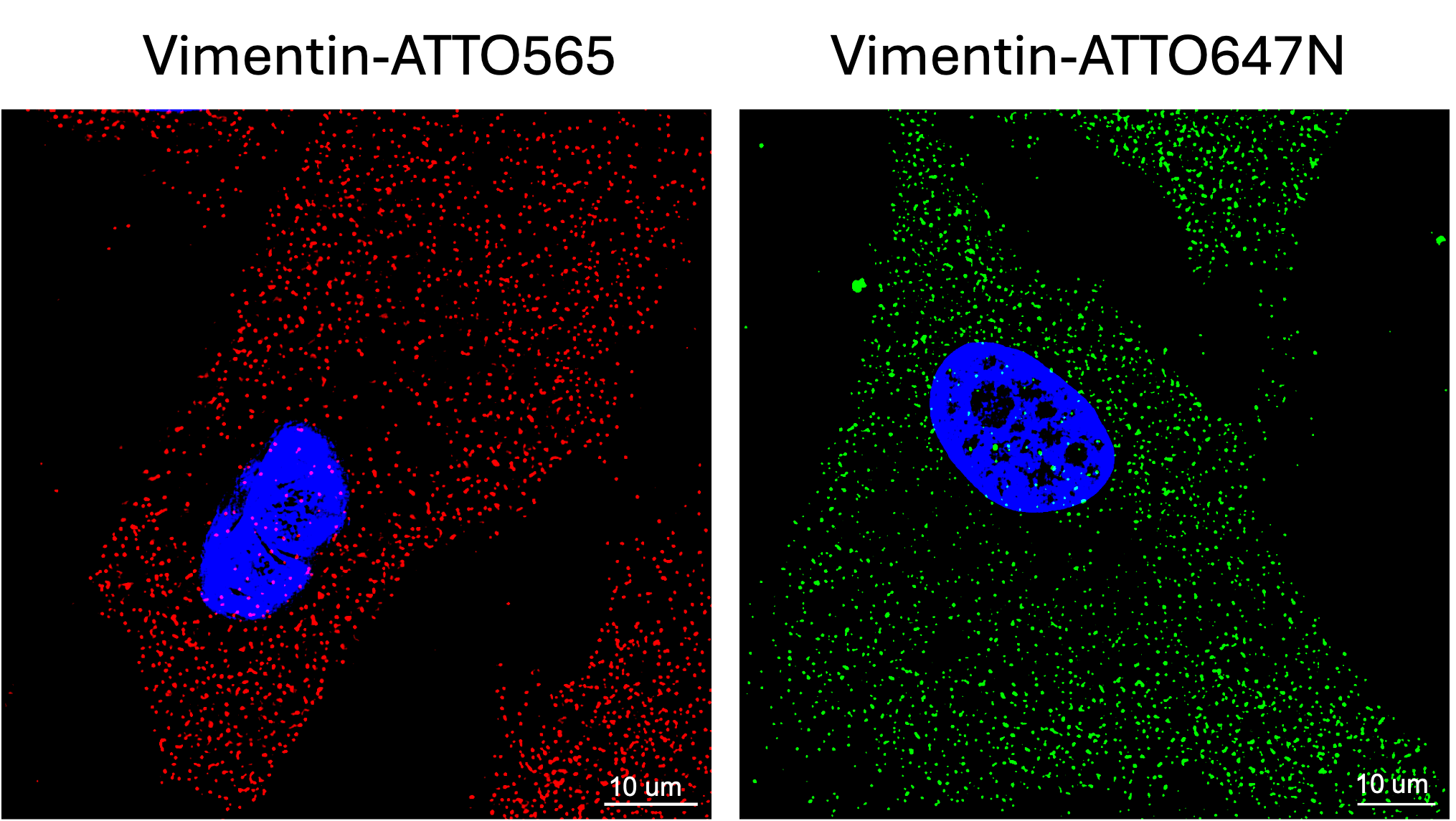
Figure 2, FISH probes labeled with a variety of fluorescent dyes: Human hepatic stellate cells (HHSC) were stained for human Vimentin mRNA with FISH probe, labeled either with ATTO565 (left, red) or ATTO647N (Right, green) dyes. Nuclei were counterstained with DAPI. Imaging was conducted using Leica TIRF/D STORM/THUNDER (widefield) with a 63x objective with oil (Right). Scale bar: 10 µm
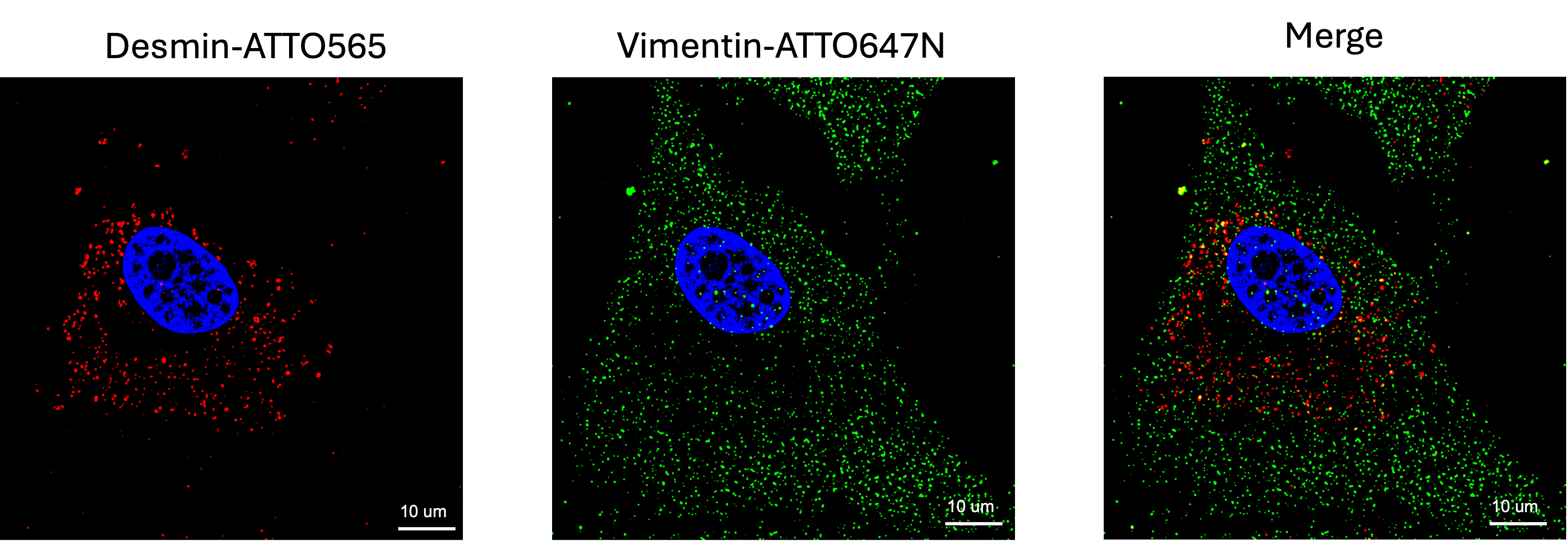
Figure 3, Dual-target probes reveal spatial distribution of mRNA: Human hepatic stellate cells (HHSC) were stained for desmin and Vimentin mRNAs with FISH probes, labeled either with ATTO565 (red) or ATTO647N (green) dyes. Nuclei were counterstained with DAPI. Imaging was conducted using Leica TIRF/D STORM/THUNDER (widefield) with a 63x objective with oil (Right). Scale bar: 10 µm
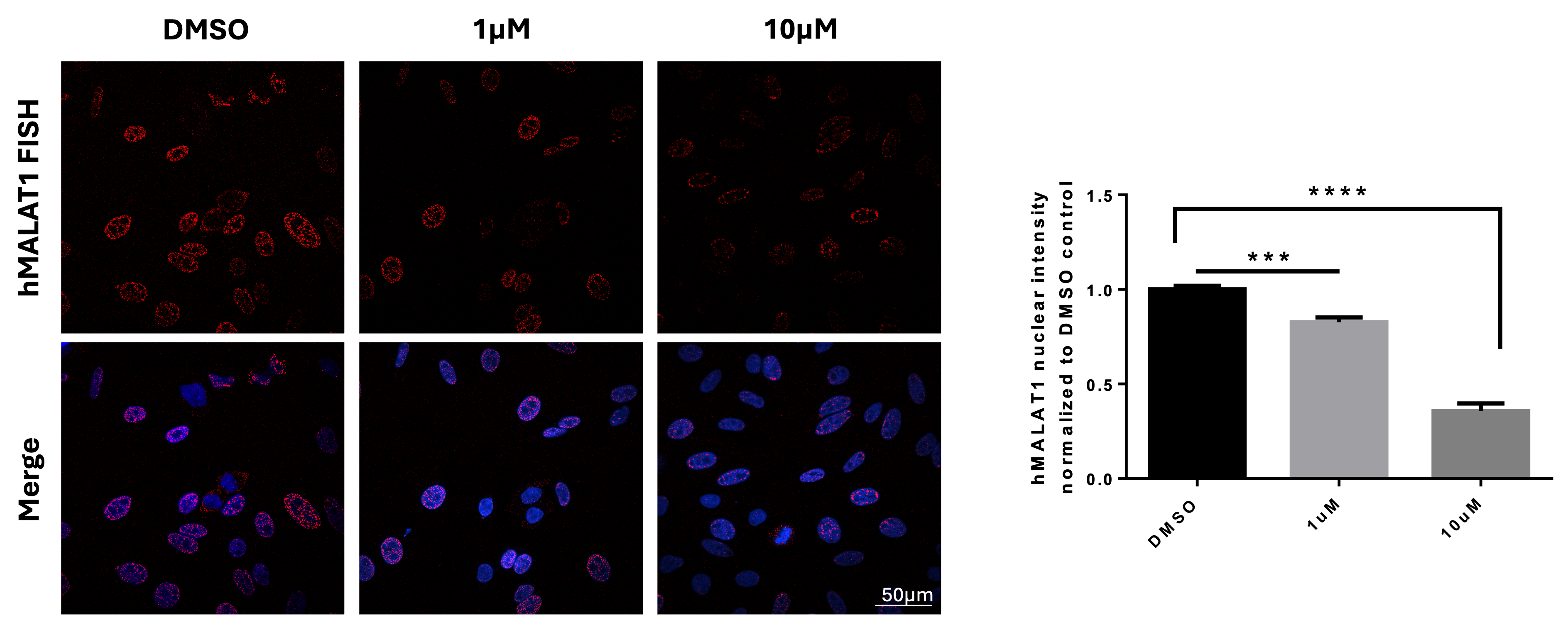
Figure 4, High-throughput FISH screening identifing Tyrphostin A9 modulating lncRNA MALAT1 in Hela cells: Hela cells were seeded in PhenoPlate 384-well black plate for overnight before staining for MALAT1 lncRNA by FISH after treatment with DMSO, 1uM, or 10uM Tyrphostin A9 for 2h. Nuclei are stained by DAPI (Right). Imaging was acquired using the automated confocal microscopy system of Nikon Eclipse Ti2-E with 60x water objective. Quantification of nuclear MALAT1 staining intensity normalized to DMSO (Left). Highly significant effect is detected. Bars show mean with SD. **** p value < 0.0001, *** p value < 0.001


